Clinical Approaches for Understanding the Expression Levels of Pattern Recognition Receptors in Otitis Media with Effusion
Article information
Abstract
Objectives
Bacterial infections in the normally sterile environment of the middle ear cavity usually trigger host immune response, whereby the innate immune system plays a dominant role as the host's first line of defense. We evaluated the expression levels of Toll-like receptors (TLRs) -2, -4, -5, -9, and nucleotide-binding oligomerization domain-containing proteins (NODs) -1 and -2, all of which are related to bacterial infection in pediatric patients with otitis media with effusion (OME).
Methods
The study sample consisted of 46 pediatric patients with OME, all of whom had ventilation tubes inserted. The expression levels of TLR-2, -4, -5, -9, NOD-1 and -2 mRNA in middle ear effusion were assessed by polymerase chain reaction (PCR). Difference of pattern recognition receptors (PRRs) expression level by presence of bacteria, ventilation tube insertion rate, and effusion fluid character was assessed.
Results
All effusion fluid samples collected from patients with OME showed expression of TLR-2, -4, -5, -9, NOD-1, and -2 mRNA by PCR. However, we found no differences among expression levels of PRRs in relation to characteristics of exudates, presence of bacteria, or frequencies of ventilation tube insertion (P>0.05).
Conclusion
Our findings suggest that exudates of OME patients show PRR expressions that are related to the innate immune response regardless of the characteristics of effusion fluid, presence of bacteria in exudates, or frequency of ventilation tube insertion.
INTRODUCTION
Otitis media with effusion (OME) is associated with bacterial infections of the upper respiratory tract and dysfunction of the Eustachian tubes. Several other factors may also be involved in the development of OME, including the host immune system, environmental factors, family history, allergies, adenoid hypertrophy, chronic sinusitis, cleft palate, tumors, or even sharp changes in atmospheric pressure. Recently, it was reported that in many cases of OME, bacteria or viruses are found in middle ear effusion. Since pediatric patients with OME often have previous histories of acute otitis media (AOM); OME is often regarded as the same disease as AOM (1). Therefore, understanding the infections involved in AOM and following the immune response in the middle ear cavity are critical to determining the pathophysiological mechanism of OME.
The human body is constantly threatened by pathogens such as viruses, bacteria, fungi and parasites. In cases of infection the innate or adaptive immune system, or a cooperative interaction of both, release various immunological mediators. The innate immune system is considered the first line of defense during the host response to pathogens, and the body discriminates and infectious non-self from noninfectious self based on the recognition of general patterns (2, 3). Pattern recognition receptors (PRRs) in humans, including Toll-like receptors (TLRs) and cytoplasmic nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), recognize conserved molecular signatures known as pathogen-associated molecular patterns (PAMPs) to sense the presence of microbial infections (4).
Recent studies on PRR expressions in pediatric patients with OME have reported that the decreased expression of PRRs may be associated with increased susceptibility to OME (5, 6). Even though no mutations had been found in TLR-2 and -4, which were expressed in all isolated samples of middle ear fluids, levels of TLR-9 and NOD-1 mRNAs were significantly lower in individuals with recurrent OME (6). Based on this, we hypothesized that there may be a difference in expression level of PRRs dependent on the clinical states of OME. Thus, in the present study we focused on the innate immune response in OME patients according to the characteristics of effusion fluids, presence of bacteria in exudates, and the frequency of ventilation tube insertion. Specifically, in the innate immune response in OME, we addressed the expressions of TLR-2, -4, -5, -9, NOD-1, and -2, which may play important roles in bacterial infection.
MATERIALS AND METHODS
The study sample consisted of 46 pediatric patients who visited the Department of Otorhinolaryngology at our hospital from January 2008 to April 2010 and underwent ventilation tube insertion for chronic OME. OME was diagnosed by the presence of an amber-colored tympanic membrane on otoscopic examination and by the presence of B- or C-type tympanograms as shown by impedance audiometry. Surgery was performed on chronic OME patients who did not show improvement after 2 weeks of antibiotic treatment and in patients who, after a 2.3 month follow-up, showed progressive retraction of the eardrum or progression of hearing loss as shown by an increase in pure tone threshold.
We received prior permission with written informed consent from the patients' parents or guardians for using patient samples, and the purpose of the experiment was also explained to them. Children who were suspected of having acute otitis media, head and neck anomalies, systemic diseases, or congenital or acquired immunodeficiencies were excluded.
A tympanostomy tube was inserted after a radial shaped incision was made in the anterior inferior quadrant of the tympanic membrane. Prior to surgery, the external auditory canal was washed with potadine solution, and middle ear effusion fluid (MEEF) was collected using a collector (Xomed Treace Products, Jacksonville, FL, USA) by aseptic procedures, taking care to avoid bleeding. Each effusion fluid sample was collected using sterilized cotton swabs, added to Stuart transport medium, and used to inoculate blood agar medium and thioglycollate liquid medium. All cultures were incubated for at least 24 hours at 35℃, and the resultant bacteria were identified by Gram-staining and biochemical analyses.
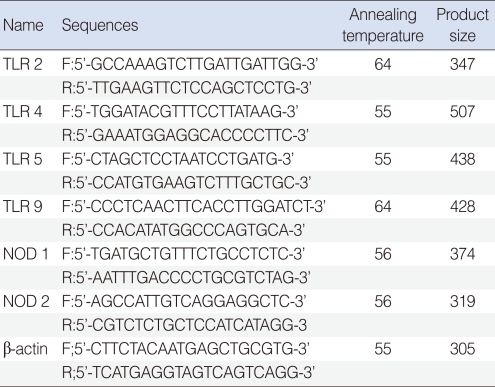
Total RNA was extracted from effusion using the RNA-Bee solution kit (Tel-Test, Friendswood, TX, USA) following the manufacturer's protocol. First-strand cDNA synthesis was performed by reverse transcription in a total volume of 20 µL reaction mixture containing 1 µg of RNA, 1x reaction buffer, 1 mM dNTP, 5 µM random primers, 20 units RNase inhibitor, and 20 units AMV reverse transcriptase (Promega, Madison, WI, USA). The reaction mixture was incubated at 42℃ for 1 hour, terminated by heating at 95℃ for 5 minutes. TLR-2, -4, -5, -9, NOD-1 and -2 primers are shown in Table 1.
Real-time polymerase chain reaction (PCR) was performed using a Chromo4 Detector real-time system (Bio-Rad, Hercules, CA, USA) with the SsoFast EvaGreen supermix (Bio-Rad). PCR was performed with 2 µL of cDNA in a 20 µL reaction mixture of 10 µL SsoFast EvaGreen supermix, 2 µL primers and 6 µL PCR grade water. The reaction parameters were as follows: denaturation at 95℃ for 30 seconds followed by 45 cycles at 95℃ for 5 seconds, 55℃ to 64℃ for 12 seconds. The crossing point of TLR-2, -4, -5, -9, NOD-1, or -2 with β-actin was applied to the formula, 2-(target gene- β actin) and the relative amounts were quantitated. Difference of PRRs expression level by presence of bacteria, ventilation tube insertion rate, and effusion fluid character was assessed.
Between group differences in PRRs expression, bacterial culture results, and frequency of ventilating tube insertion were compared using the Mann-Whitney U-test. The relationship between the expression level of PRRs and characteristics of effusion fluid was analyzed by the Kruskal Wallis test. All statistical calculations were performed using SPSS ver. 11.5 (SPSS Inc., Chicago, IL, USA), and P-values lower than 0.05 were considered statistically significant.
RESULTS
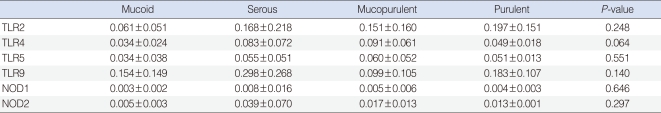
The study sample consisted of 46 children with OME, 35 boys and 11 girls with a mean age 4.7 (±2.1) years. Effusion fluids from all 46 of these patients were collected during the insertion of tympanostomy tubes. TLR-2, -4, -5, -9, NOD-1, and -2 were expressed in all middle ear exudates; TLR-9 showed the highest expression level, followed by TLR-2, TLR-4, TLR-5, NOD-2, and NOD-1. The characteristics of effusion fluid presented 17 mucoid cases, 16 serous cases, 10 mucopurulent cases and 3 purulent cases. There were no significant differences among mRNA levels of TLR-2, -4, -5, -9, NOD-1, and -2 regardless of their effusion characteristics (P>0.05) (Table 2).
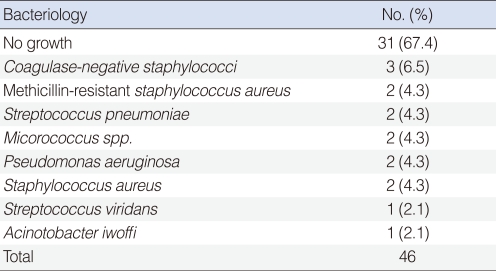
Of the 46 samples examined, 31 (67.4%) were negative for growth of bacteria, whereas the remaining 15 (32.6%) showed bacterial growth from the effusion fluid. Bacteria were cultured and identified; the bacterial species identified were coagulase-negative Staphylococcus (CNS), methicillin-resistant Staphylococcus aureus (MRSA), Streptococcus pneumonia, Micrococcus spp., Pseudomonas aeruginosa, methicillin-sensitive Staphylococcus aureus, Streptococcus viridians, and Acinotobacter iwoffi (Table 3).
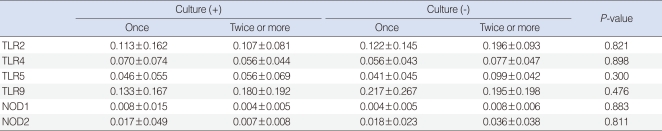
We observed expressions of TLR-2, -4, -5, -9, NOD-1, and -2 mRNAs in all effusion samples. We observed no differences between the presence of bacteria in exudates and the expression levels of TLR-9, NOD-1, and NOD-2 mRNA (P>0.05) (Table 4). In addition, expression levels of PRRs mRNA were not associated with the frequency of ventilating tube insertion (P>0.05) (Table 5). Similarly, there were no differences between frequencies of ventilation tube insertion and expression levels of PRRs, regardless of the presence of bacteria in middle ear fluid (P>0.05) (Table 6).
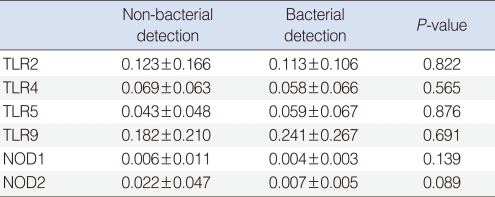
Expression of PRRs according to bacterial culture results in effusion fluid (relative expression ΔCt)
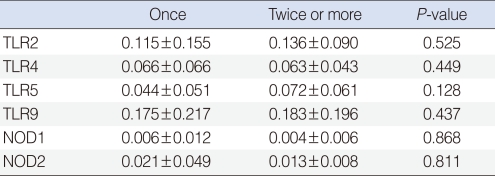
Expression of PRRs according to the frequency of ventilating tube insertion (relative expression ΔCt)
DISCUSSION
PRRs are a class of proteins that respond to PAMPs while the innate immune system reacts. These receptors are expressed in different cellular compartments such as on the cell surface, and in the endosomes, lysosomes or cytoplasm; they have been shown to differ in intracellular localization, recognition of different pathogenic motifs, and cell signaling (7). For example, TLR-2, -4, and -5 are located on the cell surface, TLR-9 is present on endosomal and lysosomal membranes, and NOD-1 and -2 receptors are located in the cytoplasm. TLR-2 and -4 have been reported to recognize pathogenic bacteria, whereas TLR-2 is the receptor for the cell wall components of Gram-positive bacteria including peptidoglycans, lipoteichoic acid and lipoproteins, and TLR-4 binds to the toxic pneumolysin ligand produced by Gram-positive bacteria, as well as binding to lipopolysaccharide (LPS), the major component of Gram-negative bacterial cell walls (8-10). Both NOD-1 and -2 have cytoplasmic caspase-recruiting effector domains and recognize peptidoglycan fragments. NOD-1 recognizes Gram-negative bacteria, whereas NOD-2 recognizes both Gram-positive and Gram-negative bacteria (11). Inflammatory diseases caused by the abnormal immune control of nosocomical infections may be due to some problem with expression levels of PRRs. Despite the fact that PRRs may play a crucial role in the innate immune response, little is known about how and when the innate immune system reacts with pathogens invading the middle ear cavity in pediatric patients with OME. Thus, we not only assayed the expressions and relationships among PRRs, but we also aimed to discern whether there were differential expression levels of PRRs according to bacterial infection.
TLRs serve a sentinel role in early detection for invading pathogens so that the host can activate local inflammation through the expression of innate immune mediators such as the complement cascade. When there is a mutation in TLRs, not only otitis media, but periodontitis and osteomyelitis can also occur due to the reduced ability for bacterial cell wall recognition and an increased susceptibility to bacterial infection (12-16). TLR-2 and -4 were reported to be involved in defense mechanism of the middle ear cavity and in the bacterial alleviation or clearance of OME. Genetic mutation of TLR-2 or -4 was also reported to increase the susceptibility for bacterial infection in OME (6, 17). In our study, we confirmed that TLR-2 and -4 were expressed in exudates of OME. TLR-5 and -9 that are related to the response to bacterial infection were also expressed in the middle ear effusion. These results imply that the difference of PRRs expression levels may be closely associated with the development of OME.
The role of cytoplasmic receptors becomes critical when pathogens that have the ability to escape from TLR recognition enter the body (18). The expression levels of many cytoplasmic receptors stay low under normal conditions, but when pathogens invade and pro-inflammatory cytokines are manifested, their expressions are highly induced. Our results showed simultaneous expression of the cytoplasmic receptors NOD-1 or NOD-2 along with the TLRs in all cases regardless of bacterial presence or species. This suggests that PRRs function cooperatively rather than individually in OME.
Previous studies on the immunological status of the host in otitis media have asserted that the independent local immunity of the middle ear cavity is one of the main causes of OME (19, 20). The close relationship between bacterial infection and the innate immune response of OME can be deduced because bacteria were detected in exudates of the middle ear cavity, and children with a higher prevalence for upper airway infection were more likely to develop OME. These findings suggest that bacterial infection probably plays an important role in the development of OME (21).
Some have presented an association between the level of antibody in the middle ear effusions and bacterial infection, and suggested that the higher concentrations of immunoglobulin in middle ear effusions from patients with acute otitis media are associated with lower detection rates of bacteria. Other research has showed that the serum levels of immunoglobulin from children with OME may be closely related to bacterial infection (20). These studies led to the examination of the relationship between bacterial infection and the acquired immune response in otitis media (1, 20). However, there was no correlation between effusion Ig concentration and the presence of bacteria in OME. In order to determine the relationship between bacterial infection and innate immunity, the expression levels of PRRs were compared between subjects who were detected of bacteria and those who were not detected of bacteria in exudates, and we found no relationship between bacterial infection and PRRs. However, it is still important to note the possible involvement of PRRs in immune response to otitis media, since PRRs were expressed in all exudate samples from otitis media regardless of bacterial detection.
There are some limitations in this study. First, the study did not include children with early stage OME. Second, antibiotics had been used for 2 weeks to treat early stage of symptoms in all patients. Third, 67.4% showed negative growth despite bacterial infection possibly due to the use of various antibiotics prior to myringotomy and insertion of ventilation tubes, creating a bacteriostatic condition and delaying the proliferation and growth of pathogens. Fourth, only the middle ear effusion was tested for bacteria and not the mucous membrane of the middle ear. The exudate contains only a few partially exfoliated mucosal epithelial cells and inflammatory cells; this would not fully reflect the immune cells in the middle ear mucosa during infection, but we obviously could not collect the middle ear mucosa of patients due to ethical issues. Finally, as the exudates used in this study were collected during surgery 2-3 months after the occurrence of the initial onset of otitis media, our study may not fully reflect the innate immune response in the middle ear cavity. PRRs were expressed in all exudates samples of otitis media regardless of bacterial detection. Our study showed similar PRRs expression values between the group that received reoperation from the recurrence of OME and the group with only one operation. This suggests that innate immunity may be involved in the middle ear cavity irrespective of the recurrence of OME.
This study confirmed that all exudates of OME patients showed some level of PRR expression related to innate immune response regardless of characteristics of effusion fluid, presence of bacteria in exudates, or frequency of ventilation tube insertion.
Notes
No potential conflict of interest relevant to this article was reported.