MicroRNA-145-5p Regulates the Epithelial-Mesenchymal Transition in Nasal Polyps by Targeting Smad3
Article information
Abstract
Objectives.
The annual prevalence of chronic rhinosinusitis (CRS) is increasing, and the lack of effective treatments imposes a substantial burden on both patients and society. The formation of nasal polyps in patients with CRS is closely related to tissue remodeling, which is largely driven by the epithelial-mesenchymal transition (EMT). MicroRNA (miRNA) plays a pivotal role in the pathogenesis of numerous diseases through the miRNA-mRNA regulatory network; however, the specific mechanism of the miRNAs involved in the formation of nasal polyps remains unclear.
Methods.
The expression of EMT markers and Smad3 were detected using western blots, quantitative real-time polymerase chain reaction, and immunohistochemical and immunofluorescence staining. Differentially expressed genes in nasal polyps and normal tissues were screened through the Gene Expression Omnibus database. To predict the target genes of miR-145-5p, three different miRNA target prediction databases were used. The migratory ability of cells was evaluated using cell migration assay and wound healing assays.
Results.
miR-145-5p was associated with the EMT process and was significantly downregulated in nasal polyp tissues. In vitro experiments revealed that the downregulation of miR-145-5p promoted EMT. Conversely, increasing miR-145-5p levels reversed the EMT induced by transforming growth factor-β1. Bioinformatics analysis suggested that miR-145-5p targets Smad3. Subsequent experiments confirmed that miR-145-5p inhibits Smad3 expression.
Conclusion.
Overall, miR-145-5p is a promising target to inhibit nasal polyp formation, and the findings of this study provide a theoretical basis for nanoparticle-mediated miR-145-5p delivery for the treatment of nasal polyps.
INTRODUCTION
Chronic rhinosinusitis (CRS) is an inflammatory condition affecting the nasal mucosa and sinuses [1]. It is classified into chronic rhinosinusitis with nasal polyps (CRSwNP) and chronic rhinosinusitis without nasal polyps based on the presence or absence of nasal polyps [2]. The manifestations of CRS encompass nasal congestion, runny nose, maxillofacial pain, impaired olfactory perception, and even anosmia [3]. Nasal polyps are inflammatory lesions that originate in the mucous membranes of the sinuses and can protrude into the nasal passages or be found in the sinuses themselves. The etiology of nasal polyps is thought to be closely related to dysfunction of the nasal mucosal epithelial cells, microbial and bacterial infections, and disturbances in the host’s immune system [4], although the exact mechanisms are not fully understood. The incidence of CRSwNP has been increasing in recent years. The primary treatment options include surgical procedures and medication, both of which can significantly affect patients’ quality of life due to their limited efficacy [5]. Simultaneously, this disease imposes a substantial economic burden upon society [6]. Therefore, it is imperative to study the mechanism underlying the formation or development of nasal polyps.
The epithelial-mesenchymal transition (EMT) is a dynamic and reversible differentiation process, wherein epithelial cells lose their characteristic attributes and progressively acquire mesenchymal traits, transforming into mesenchymal cells guided by inducible transcription factors [7,8]. The EMT is a critical part of cell growth and development, primarily contributing to embryonic and organ development, wound healing, tissue regeneration and remodeling, and fibrosis [9], and it is also associated with tumorigenesis [10]. Abnormal EMT has been linked to the development of various diseases, including retinal dysfunction, proliferative scarring, keloid formation, and respiratory organ fibrosis [11-13]. Prior research has demonstrated that the EMT plays a key role in nasal polyp formation. Studies have shown that the reduction of the EMT in the nasal mucosal epithelium can lead to a decrease in nasal polyps [14,15]. The EMT is recognized as a key factor in the complex remodeling of nasal mucosal tissue and in sustaining the inflammatory environment within the nasal cavity [16]. Nevertheless, the exact mechanisms governing this phenomenon remain unknown.
MicroRNAs (miRNAs) are short RNA molecules of approximately 19–25 nucleotides [17] that modulate gene expression through their interaction with the 3’-untranslated region (UTR) of particular target mRNAs, and are involved in apoptosis, proliferation, differentiation, and regeneration of cells [17,18]. Previous studies have found aberrant miRNA expression in a range of diseases, such as endometriosis and tumor resistance [19-21]. In recent years, as our understanding of miRNAs has grown, they have emerged as novel therapeutic approaches for various diseases [22] and have also been used as diagnostic and prognostic biomarkers for many cancers [23]. Studies have shown that delivering miRNAs via nanoparticles is not only effective in halting the progression of diseases such as cancer, neurodegenerative disorders, and in tissue regeneration, but it also results in fewer side effects than other treatment methods [24]. Thus, miRNA-based therapies have great potential. miRNAs have been closely linked to the EMT process. They have been found to promote skin inflammatory responses in patients with psoriasis by engaging in the Wnt/β-catenin pathway [25] and to encourage the formation of wound scar tissue through the transforming growth factor (TGF)-β pathway [26]. It is recognized that the Wnt/β-catenin pathway and the TGF-β pathway are closely related to the EMT [8]. Additionally, the formation of nasal polyps may be associated with a variety of miRNAs, but the key miRNAs and the mechanisms are still unknown. Therefore, identifying these crucial miRNAs and elucidating the mechanisms by which they contribute to nasal polyp formation is of paramount importance.
In this study, we determined that miR-145-5p exhibits different levels of expression in nasal polyp tissues compared to normal tissues. Further investigation revealed that the upregulation of miR-145-5p can mitigate the EMT induced by TGF-β1. Mechanistically, the upregulation of miR-145-5p suppresses EMT by reducing the expression of Smad3. Consequently, miR-145-5p is anticipated to be a crucial target for nasal polyp treatment.
MATERIALS AND METHODS
miRNA expression data analysis
Expression profiling data of gene array GSE169376, encompassing six nasal polyp tissues and three normal tissues, were acquired from the Gene Expression Omnibus (GEO) database [27]. The miRDB, miRTarBase, and TargetScan databases were enlisted to forecast the potential target sites of miR-145-5p. The entire dataset was analyzed within the statistical environment of the R language. The R language version is 4.2.2, and the packages used are library (GEOquery), library (limma) and library (umap). For further analysis and visualization, the online resources OmicShare (www.omicshare.com/tools) and miRPath were employed.
Patients
In this study we collected inferior turbinate tissues from 8 control patients and nasal polyp tissues from 15 patients with CRSwNP; 17 patients were males and 6 patients were females; the average age of the patients in the control group was 32 years old, and the average age of the patients in the experimental group was 44 years old. Nasal polyp tissues were taken from the middle nasal passage region, while normal tissue was procured from patients with inferior turbinate hypertrophy but devoid of any sinus disease. The study excluded patients with acute infections, fungal sinusitis, and cystic fibrosis as per the diagnostic criteria recommended by the European Position Paper Guidelines on Rhinosinusitis and Nasal Polyps [28]. The exclusion criteria also included pregnant or lactating women, individuals with malignancies, and hematologic disorders. The patients were evaluated based on their medical history and examination to ensure compliance with the criteria [28]. None of the patients had received corticosteroids, antibiotics, or antihistamines in the 4 weeks before surgery. All specimen tissues were partitioned into three segments: one was preserved in RNAlater to facilitate RNA extraction, another was fixed in 4% paraformaldehyde (PFA) to facilitate immunohistochemical staining, and the final segment was stored at –80 °C for western blot analysis.
This study received approval from the Human Ethics Committee (No. 2023-336), and all patients provided written informed consent.
Cell culture
Primary human nasal epithelial cells (HNEpCs) were purchased from Suzhou Baiske Biotechnology Co. HNEpCs were cultured in EMEM (Eagle’s Minimum Essential Medium) containing 10% fetal bovine serum (FBS; Gibco). Then, the cells were placed in a 5% CO2 chamber at 37 °C for incubation. HNEpCs were seeded into 25 cm2 culture flasks (T25 flask) and employed for subsequent experiments. The EMT model was established by employing recombinant human TGF-β1 (HY-P7118, MedChemExpress). Synthesize miRNA mimic (B01001, GenePharma), miRNA inhibitor (B03001, GenePharma), and construct Smad3 lentiviral (D02001, GenePharma) transfected HNEpCs. Transfection was conducted following the manufacturer’s provided guidelines.
Immunofluorescence staining
HNEpCs were placed on sterile slides and cultured within 24-well culture plates. Subsequently, they were rinsed with phosphate-buffered saline (PBS) for 5 minutes, followed by fixation with 4% PFA for precisely 15 minutes, and permeabilizated with 0.2% Triton X-100 and blocking with QuickBlock blocking buffer, after which they were incubated with the primary antibody for an extended period of 12 hours at a temperature of 4 °C. Then, the slides were washed with PBS, and the cells were cultured with a fluorescently tagged secondary antibody for 1 hour at the room’s ambient temperature. The cells were observed with a fluorescence microscope (Zeiss).
Quantitative real-time polymerase chain reaction
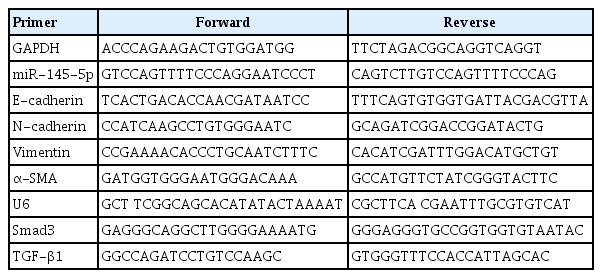
According to the manufacturer’s guidelines, TRIzol reagent was employed to extract total RNA, 1 μg of RNA was reverse transcribed into cDNA for amplification using the All-in-one cDNA synthesis kit (Yeasen), and quantitative real-time polymerase chain reaction (qRT-PCR) amplification was performed using SYBR Green PCR Master Mix. RNA integrity and the efficacy of the reverse transcription reaction were assessed through qRT-PCR amplification of glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Genewiz) or U6 (Genewiz) transcripts. The levels of miRNAs and mRNAs were normalized utilizing U6 and GAPDH, respectively, and the relative mRNA levels of the target genes were scrutinized using the 2–ΔΔCT method. The primer sequences are shown in the Table 1, including E-cadherin (E-cad; Genewiz), N-cadherin (N-cad; Genewiz), Vimentin (Vim; Genewiz), α-smooth muscle actin (α-SMA; Genewiz), and TGF-β1 (Genewiz).
Western blot analysis
The gathered tissues were pulverized in liquid nitrogen. Radioimmunoprecipitation assay lysis buffer with a protease inhibitor mixture was used to extract total proteins. The protein concentration in the supernatant was evaluated by employing the BCA assay kit, and thereafter, the supernatant was subjected to boiling for 10 minutes while being mixed with protein uploading buffer. Sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was used to segregate proteins, and then the proteins were transferred to polyvinylidene fluoride membranes. Membranes were enclosed with a 5% skimmed milk solution in Tris-buffered saline Tween (TBST) buffer, which also contained 0.05% Tween, and were incubated at ambient temperature for 1 hour. Following this, primary antibodies against E-cad (A22333, ABclonal), N-cad (A21308, ABclonal), Vim (A19607, ABclonal), TGF-β1 (A2124, ABclonal), Smad3 (A19115, ABclonal), and p-Smad3 (AF3362, Affinity Biosciences) were incubated overnight at 4 °C. The primary antibodies were diluted to a concentration of 1:1,000. At room temperature, the membrane was washed with TBST three or four times for 10 minutes, and then incubated with the secondary antibody (A0208, Beyotime) for 1 hour at ambient temperature. The secondary antibody was diluted to a concentration of 1:5,000. Ultimately, beheld the immunoreactivity through the enhanced chemiluminescence kit. The imaging system was used to ascertain the intensity of the protein bands.
Histological analysis
The gathered nasal polyp tissues and inferior turbinate tissues were dehydrated, then embedded in paraffin, after that sectioned into slices, and poised on slides for the immunohistochemical staining, employing the peroxidase-labeled streptavidin-biotin technique. The sections were immersed overnight at a temperature of 4 °C, followed by the presence of primary antibodies directed against E-cad, N-cad, Vim, and Smad3, each at dilution of 1:200. Then, the sections were incubated with the horseradish peroxidase-labeled secondary antibody on the subsequent day. All samples were visualized by the diaminobenzidine test kit and sections were counterstained with 10% hematoxylin. Protein expression levels were observed by Zeiss light microscopy, and the final evaluation of staining was performed by staining intensity and distribution.
Wound-healing assay
Cells were seeded into 6-well plates. Subsequently, the cells were treated with TGF-β1, miR-145-5p inhibitor, or miR-145-5p mimic. Upon reaching a state with a cell density of 100%, the monolayer cell planes were scraped using the tip of a sterilized 200 µL pipette. The resultant trauma scratches were observed and recorded with a light microscope (Zeiss).
Cell migration assay
The cells were resuspended in EMEM without FBS. Subsequently, cells were seeded into the 24-well plate. Then 500 µL of EMEM containing 20% FBS was added in the lower chamber as an inducer of the cell. Following the incubation period, the migrating cells were immobilized with a 4% PFA solution for half an hour, followed by staining with 5% crystal violet. Migrated cells were observed and recorded by an inverted microscope (Zeiss).
Luciferase reporter assay
Cells were inoculated into 12-well plates. Wild-type (WT; C09005) and mutant (Mut; C09006) luciferase reporter plasmids (GenePharma) were constructed. miR-145-5p-mimic (B02001, GenePharma) or NC-mimic (B02001, GenePharma) and plasmids were transfected into cells simultaneously. Forty-eight hours after transfection, cells were lysed and firefly luciferase activity was measured with a Dual-Luciferase Reporter Assay System (Promega).
Statistical analysis
GraphPad Prism software version 8.0 (GraphPad Software) was used to analyze for all quantitative data, which were displayed as the mean±standard deviation. Statistical significance was determined by one-way or two-way analysis of variance with Tukey’s test, with a P-value <0.05 regarded as significant. At least three independent experiments contributed to the findings of this study.
RESULTS
Altered expression of EMT markers in nasal polyp tissues
To determine whether the EMT occurred in nasal polyp tissues, the expression of EMT markers was examined in nasal polyps and normal tissues. Nasal polyp tissues and inferior turbinate tissues were subjected to qRT-PCR, western blotting, and immunohistochemical staining experiments (Fig. 1A). Epithelial markers including E-cad, N-cad, Vim, and α-SMA were predominantly expressed within the mesenchymal tissue. Western blot analysis showed that the expression of E-cad was lower than in normal tissues (Fig. 1B-E). In contrast, the expression of N-cad and Vim was significantly higher than in normal tissues (Fig. 1B-E). The qRT-PCR results displayed elevated expression levels of Ncad, Vim, and α-SMA compared with normal tissues (Fig. 1F-H). Subsequently, the localization of the markers and their differential expression were assessed by immunohistochemical staining, and E-cad and N-cad were discernible in the nasal mucosa and glandular epithelium, respectively, while Vim was present in the submucosal and perivascular regions (Fig. 1I). Notably, the expression of E-cad decreased in nasal polyp tissues, whereas the expression of N-cad and Vim increased (Fig. 1I), which aligned with the findings from qRT-PCR and western blotting. In addition, qRT-PCR and western blotting were used to detect TGF-β1 in normal tissues and nasal polyp tissues, and the results showed that the expression of TGF-β1 was higher in nasal polyp tissues (Supplementary Fig. 1). These results suggested that EMT was present in nasal polyp tissues.

Expression of epithelial-mesenchymal transition markers in nasal polyp tissue. (A) Nasal structure and scheme of the study design. (B-E) Western blot analysis of protein expression in normal tissues (N1, N2, N3) and nasal polyps (P1, P2, P3). ImageJ was used to calculate the relative expression rate. (F-H) Quantitative real-time polymerase chain reaction (qRT-PCR) for mRNA expression in normal tissues and nasal polyp tissues. (I) Representative images of immunohistochemical staining in normal tissues and nasal polyp tissues. Scale bar=100 μm (left) and 20 μm (right). All data are shown as mean±standard deviation. n=3 per group. SMA, smooth muscle actin. *P<0.05, **P<0.01, as compared to the control group (normal tissues).
miR-145-5p was downregulated in nasal polyp tissues
The GSE169376 dataset was analyzed using the GEO2R software available in the GEO database. In total, 23 miRNAs were identified as differentially expressed miRNAs between nasal polyps and normal tissues based on the screening criteria of a P<0.05, an adjusted P<0.05, and an absolute logFC value >1, and all of these miRNAs exhibited low expression in nasal polyp tissues (Fig. 2A). miR-145-5p, miR-30a-3p, miR-27b-3p, miR-23a-5p, miR-23b-3p, miR-132-3p, and miR-193b-5p were associated with the TGF-β pathway according to Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis (Fig. 2B). miR-145-5p was involved not only in the TGF-β signaling pathway, but also in processes such as bacterial invasion of epithelial cells, adherens junctions, and the PI3K-Akt signaling pathway by KEGG enrichment analysis (Fig. 2C). These pathways were associated with the EMT. Next, we corroborated the findings through qRT-PCR analysis, which indicated a notably diminished expression level of miR-145-5p in nasal polyp tissues compared to normal tissues (Fig. 2D-J). These findings indicated that miR-145-5p was significantly downregulated in nasal polyp tissues and associated with the EMT.
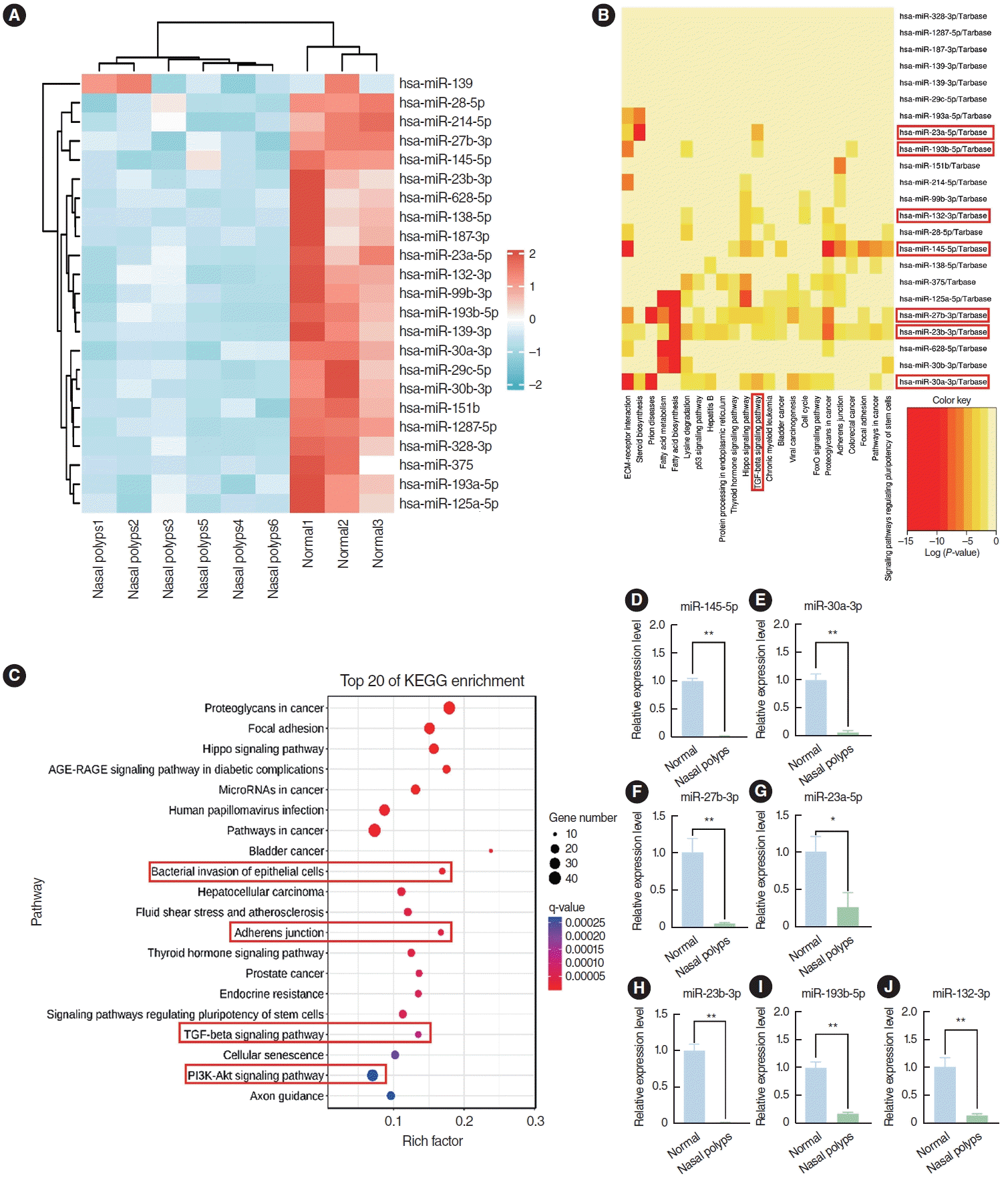
miR-145-5p was significantly low-expressed in nasal polyp tissues. (A) Heatmap of the differential expression of genes between nasal polyp tissues and normal tissues. The red hue signifies elevated gene expression, whereas the blue shade corresponds to diminished gene expression. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of miRNAs. Darker hues indicate a more robust correlation. (C) KEGG enrichment analysis of miR-145-5p. (D-J) The expression levels of miRNAs in normal and nasal polyp tissues were verified through quantitative real-time polymerase chain reaction. The levels of miRNAs were normalized utilizing U6. All data are shown as mean±standard deviation. n = 3 per group. *P<0.05, **P<0.01.
TGF-β1 induced the EMT and inhibited the expression of miR-145-5p
To investigate the optimal concentration and time of TGF-β1 in inducing EMT in HNEpCs, we conducted a stimulation experiment using TGF-β1 at concentrations of 0, 5, and 10 ng/mL, while keeping the time of stimulation constant. Additionally, HNEpCs were also subjected to TGF-β1 treatment at the same concentration for periods of 0, 12, 24, and 48 hours. A light microscope was then used to observe the cell morphology (Fig. 3A). The findings indicated that, over time and with increasing concentration, the expression of E-cad gradually decreased, while the expression of N-cad and Vim progressively increased (Fig. 3B-I). This trend was most pronounced at a concentration of 10 ng/mL and after 48 hours. At a concentration of 10 ng/mL, after treatment for 48 hours, the morphology of the HNEpCs underwent a transformation, transitioning from a cobblestone-like arrangement to an elongated, spindle-shaped fibroblast morphology (Fig. 3J). Based on this, we opted to induce cells with a concentration of 10 ng/mL TGF-β1 for 48 hours to establish the EMT model for the forthcoming experiment. Subsequently, we assessed the mRNA expression of miR-145-5p, α-SMA, N-cad, and Vim, and the results obtained from qRT-PCR analysis revealed downregulated miR-145-5p expression and increased expression of α-SMA, N-cad and Vim mRNA (Fig. 3K-N). In summary, TGF-β1 induced the EMT and inhibited the expression of miR-145-5p in HNEpCs.

Transforming growth factor (TGF)-β1 induced the epithelial-mesenchymal transition (EMT) and inhibited the expression of miR-145-5p in human nasal epithelial cells (HNEpCs). (A) Experimental protocol for observing morphological changes. (B-E) Western blot analysis of the expression levels of EMT markers in HNEpCs treated with various concentrations of TGF-β1. (F-I) Western blot analysis was conducted to evaluate fluctuations in the expression levels of EMT markers in HNEpCs after varying time intervals. (J) After 48 hours with 10 ng/mL TGF-β1, changes in the cell morphology of HNEpCs. Scale bar=100 μm (left) and 20 μm (right). (K-N) The mRNA expression levels of miR-145-5p and EMT markers in HNEpCs, treated with 10 ng/mL TGF-β1 for 48 hours. All data are shown as mean±standard deviation. n = 3 per group. NS, not significant; SMA, smooth muscle actin. *P<0.05, **P<0.01.
miR-145-5p inhibited the EMT and cell migration
To elucidate the role of miR-145-5p in the EMT, we initially transfected HNEpCs with a miR-145-5p inhibitor. qRT-PCR was employed to authenticate the transfection efficiency (Fig. 4A). We examined changes mRNA expression levels through qRT-PCR, and the findings revealed an increase in the expression of N-cad, Vim, and α-SMA after the transfection of cells with the miR-145-5p-inhibitor (Fig. 4B-D). Protein expression levels were assessed through western blotting and immunofluorescence staining. The results exhibited a reduction in the expression of E-cad, while the expression of N-cad and Vim increased (Fig. 4E-H). Concurrently, immunofluorescence staining confirmed the decrease in E-cad and the increase in Vim (Fig. 4I and J). When miR-145-5p was downregulated, there was a significant increase in cell migration, as demonstrated by wound healing and cell migration assays (Fig. 4K and L). Additionally, light microscopy revealed a morphological transition in the transfected HNEpCs from a cobblestone-like appearance to an elongated, spindle-shaped, fibroblastic form (Fig. 4M).

The downregulation of miR-145-5p promoted the epithelial-mesenchymal transition and cell migration. (A) Quantitative real-time polymerase chain reaction (qRT-PCR) for verification of miR-145-5p inhibitor transfection efficiency. (B-D) qRT-PCR for quantification of α-smooth muscle actin (SMA), N-cadherin, and vimentin mRNA expression levels. (E-H) Western blot assay for evaluation of E-cadherin, N-cadherin, and vimentin protein expression. (I, J) Immunofluorescence staining analysis to assess the expression levels of E-cadherin and vimentin. Scale bar=50 μm. (K) The scratch assay was utilized to assess the migratory capacity of cells. Scale bar=100 μm. (L) Cell migration assay was employed to evaluate of cell migratory capability. Scale bar=100 μm. (M) Observation of cell morphology under a light microscope. Scale bar=100 μm. All data are shown as mean±standard deviation. n=3 per group. NS, not significant; DAPI, 4´,6-diamidino-2-phenylindole. *P<0.05, **P<0.01.
Having confirmed the capability of TGF-β1 to induce EMT, the miR-145-5p mimic was used to transfect HNEpCs that were induced by TGF-β1. qRT-PCR was employed to validate the efficacy of the miR-145-5p mimic (Fig. 5A). Through the application of western blotting and immunofluorescence staining to gauge protein expression levels, we observed an elevation of E-cad and a concurrent reduction of N-cad and Vim, in contrast to the TGF-β1 intervention group (Fig. 5B-E). Upon upregulation of miR-145-5p, light microscopy revealed a transition in the morphology of the transfected cells to a cobblestone shape (Fig. 5F). Furthermore, qRT-PCR was employed to ascertain changes in mRNA expression levels, revealing a decrease in the expression of N-cad, Vim, and α-SMA after transfection with miR-145-5p mimic, in comparison to the TGF-β1 intervention group (Fig. 5G-I). In both wound healing assays and cell migration assays, it was observed that the upregulation of miR-145-5p led to the inhibition of cell migration (Fig. 5K and L). A schematic diagram of the cell migration assay is shown in Fig. 5J. Moreover, immunofluorescence staining noted an increase in E-cad and a decrease in the expression of Vim (Fig. 5M and N). Collectively, miR-145-5p exerts an inhibitory effect on the process of EMT, concurrently suppressing cell migration.

The upregulation of miR-145-5p inhibited the epithelial-mesenchymal transition and cell migration. (A) Quantitative real-time polymerase chain reaction (qRT-PCR) for verification of miR-145-5p mimic transfection efficiency. (B-E) Western blotting for detecting the protein expression levels of E-cadherin, N-cadherin, and vimentin. (F) Observation of cellular morphological changes under the light microscope. Scale bar=100 μm. (G-I) qRT-PCR was employed to detect the mRNA expression levels of vimentin, α-smooth muscle actin (SMA), and N-cadherin. (J) Schematic diagram of the cell migration assay experiment. (K, L) Cell migration assay and scratch assays were utilized to assess the migratory capacity of cells. Scale bar=100 μm. (M, N) Immunofluorescence staining detection of E-cadherin and vimentin expression. Scale bar=50 μm. All data are shown as mean±standard deviation. n=3 per group. TGF, transforming growth factor. *P<0.05, **P<0.01.
miR-145-5p inhibited Smad3 expression by targeting the 3’UTR sequence
Bioinformatics methodologies were employed to predict miR-145-5p targets using three databases: miRDB, TargetScan, and miRTarBase. Through this analysis, six potential target candidates were identified as present in all three datasets (Fig. 6A). Upon conducting KEGG enrichment analysis, a significant association between Smad3 and the TGF-β1 pathway was observed, as well as its correlation with regulation of the EMT (Fig. 6C). By searching the TargetScan database, we determined that miR-145-5p is complementary to Smad3 (Fig. 6D). A dual-luciferase reporter assay has provided evidence of a targeting connection between miR-145-5p and Smad3 in proliferative scars and other diseases [26], and the results of the dual-luciferase reporter assay in this study were consistent with previous studies (Fig. 6B). To further clarify the relationship between miR-145-5p and Smad3, HNEpCs were transfected with a miR-145-5p inhibitor and a miR-145-5p mimic, and western blotting and qRT-PCR experiments were subsequently conducted. The western blotting results demonstrated increased expression of Smad3 following miR-145-5p downregulation (Fig. 6E and F). Additionally, in the miR-145-5p mimic group, the expression of Smad3 was lower than in the TGF-β1 intervention group (Fig. 6G and H). The findings from the qRT-PCR experiments were consistent with the outcomes of the western blot analyses (Fig. 6I and J). The immunofluorescence staining results revealed greater fluorescence intensity of Smad3 following miR-145-5p downregulation, and the upregulation of miR-145-5p after TGF-β1 treatment led to diminished fluorescence intensity of Smad3 when compared to the TGF-β1 group (Fig. 6K). In summary, we determined that miR-145-5p exhibits a targeting relationship with Smad3, resulting in the inhibition of Smad3 expression.

miR-145-5p inhibited Smad3 expression by targeting the 3’-untranslated region (UTR) sequence. (A) Candidate genes targeted by miR-145-5p were screened using a Venn diagram. (B) A luciferase reporter assay verified the binding between miR-145-5p and Smad3 mRNA 3’-UTR. (C) Kyoto Encyclopedia of Genes and Genomes enrichment analysis was conducted on the six candidate target genes. (D) miR-145-5p was identified to have a complementary relationship with Smad3 through the TargetScan database. (E-H) Western blot analysis examining changes in Smad3 expression levels. (I, J) Quantitative real-time polymerase chain reaction was utilized to detect the mRNA expression levels of Smad3. (K) Immunofluorescence staining was used to detect alterations in the expression of Smad3. Scale bar=50 μm. All data are shown as mean±standard deviation. n=3 per group. NC, negative control; WT, wild-type; TGF, transforming growth factor. *P<0.05, **P<0.01.
miR-145-5p inhibited the EMT by directly downregulating Smad3
We investigated the expression of Smad3 and p-Smad3 in nasal polyp tissues using western blotting, qRT-PCR, and immunohistochemical staining. The results indicated that the expression of Smad3 and p-Smad3 in nasal polyp tissues were elevated in comparison to that in normal tissues, and the extent of Smad3 phosphorylation was elevated in the nasal polyps (Fig. 7A-D). Subsequently, rescue experiments were performed to validate whether miR-145-5p inhibits the EMT in cells through the direct downregulation of Smad3. qRT-PCR and Western blotting were utilized to validate the transfection efficiency (Fig. 7E-G). In comparison to the miR-145-5p mimic transfection group, upon overexpression of Smad3, the qRT-PCR results demonstrated elevated levels of N-cad, Vim, and α-SMA (Fig. 7H-J). Additionally, western blot analysis indicated a decrease in the expression of E-cad and an increase in the expressions of N-cad and Vim (Fig. 7K-N). The upregulation of Smad3 reinstated the inhibitory impact of elevated miR-145-5p on Smad3. Hence, we proposed that the downregulation of miR-145-5p increased the expression of Smad3, thereby contributing to the development of nasal polyps by promoting the EMT (Fig. 7O). Conversely, the overexpression of miR-145-5p hinders the EMT process by diminishing the expression of Smad3. These results indicated that miR-145-5p inhibited the EMT by directly downregulating Smad3.
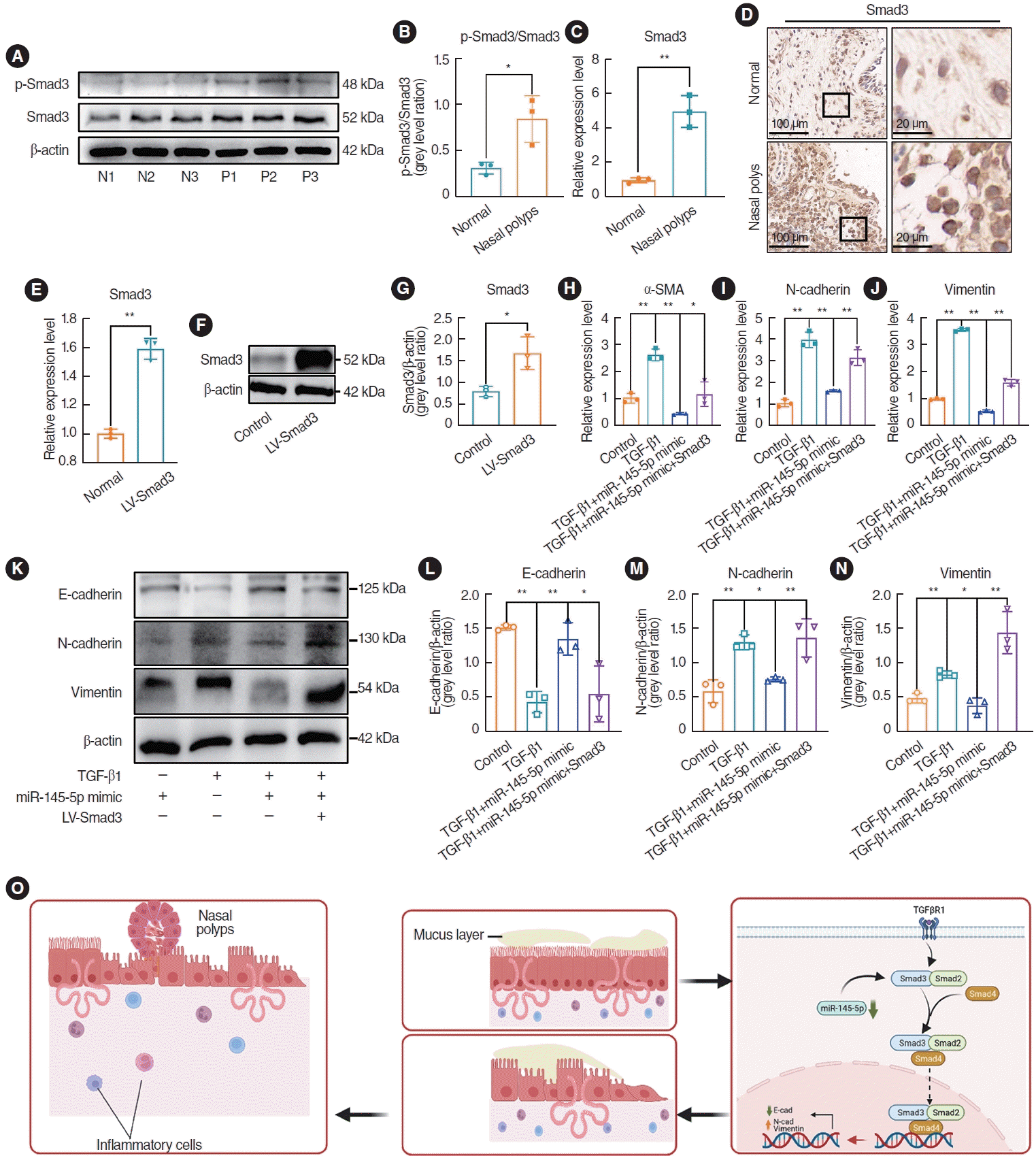
miR-145-5p exerted its inhibitory effect on the epithelial-mesenchymal transition by directly downregulating Smad3. (A, B) Western blotting was used to detect Smad3 and p-Smad3 expression in nasal polyp tissues and normal tissues. (C, D) Quantitative real-time polymerase chain reaction (qRT-PCR) and representative images of immunohistochemical staining were used to detect Smad3 expression in nasal polyp tissues and normal tissues. Scale bar=100 μm (left) and 20 μm (right). (E) Assessment of Smad3 transfection efficiency through qRT-PCR. (F, G) Assessment of Smad3 transfection efficiency through Western blotting. (H-J) qRT-PCR was employed to assess the mRNA expression of α-smooth muscle actin (SMA), N-cadherin, and vimentin. (K-N) Western blotting was employed to assess the expression of E-cadherin, N-cadherin, and vimentin. (O) Diagram of the mechanism of nasal polyp formation. All data are shown as mean±standard deviation. n = 3 per group. LV, lentivirus vector; TGF, transforming growth factor. *P<0.05, **P<0.01.
DISCUSSION
Bacteria, viruses, and inflammatory cytokines disrupt the integrity of the epithelial barrier, instigating a cascade of inflammatory responses and infections. Inflammatory responses and infections lead to the accumulation of inflammatory cells, among which macrophages and epithelial cells produce TGF-β1 [29]. TGF-β1 contributes to progressive epithelial fibrosis and tissue remodeling, and is associated with the EMT [30].
In recent years, mounting evidence has highlighted the importance of tissue remodeling induced by the EMT in the process of nasal polypogenesis. In the current study, we similarly found that the EMT is involved in the formation of nasal polyps by detecting markers of the EMT in nasal polyp tissues. Zhong et al. [31] observed hypoxic conditions influenced the differentiation of nasal epithelial cells and increased fibroblast proliferation. Furthermore, hypoxia elicited an upregulation of α-SMA and concurrent downregulation of E-cad [32]. This aligns with our study. In our investigation, we observed that nasal mucosal epithelial cells undergoing the EMT not only exhibited a downregulation of E-cad and an upregulation of α-SMA, but also displayed alterations in cell morphology. Moreover, a trend toward upregulation was noted in N-cad and Vim.
miRNAs exhibit a remarkable regulatory capacity and are implicated in a wide range of human diseases [33]. Extensive research has revealed a close link between miRNAs and the EMT [34]. In this study, we conducted a bioinformatics analysis and identified several miRNAs associated with the EMT, including miR-145-5p. In chronic obstructive pulmonary disease (COPD), it was found that low expression of miR-145-5p promoted DUSP6 expression, leading to airway remodeling [35]. Low expression of miR-145-5p was similarly found to cause keloid formation in skin diseases [36]. Airway remodeling in COPD and keloid formation in skin diseases were both associated with the EMT [37]. Similarly, the present study found that low expression of miR-145-5p was associated with nasal polyp formation. In addition to miR-145-5p, previous studies had reported that multiple miRNAs were all involved in the EMT by targeting different targets. miR-214 was overexpressed in CRSwNP and regulated the inflammatory response in CRSwNP by directly targeting SIRT1 [38]. The mechanism by which miR-21, which is overexpressed in CRSwNP, is involved in nasal polyp formation may be related to the TGF-β1-miR-21-PTEN-Akt axis [39]. In contrast, miR-761 was expressed at low levels in CRS, and overexpressed miR-761 inhibited nasal mucosal remodeling by inhibiting its target LCN2 [40]. In contrast to the above miRNAs, we identified that miR-145-5p modulates the EMT by targeting Smad3, participating in nasal polyp formation.
Silencing miR-145-5p induced the EMT in HNEpCs. Intriguingly, transfecting a miR-145-5p mimic into TGF-β1-induced HNEpCs triggered a rise in downregulated epithelial markers and a decline in elevated mesenchymal markers, signifying a reversal of the EMT. Nonetheless, our experimental outcomes indicated that the upregulation of miR-145-5p failed to comprehensively reverse the EMT. The possible reasons for this limitation are as follows. First, the dysregulated expression of multiple miRNAs leads to the EMT. Second, the pathways involved in the EMT include the nuclear factor kappa B, Wnt/β-catenin, and Akt pathways apart from the well-known TGF-β pathway [8,41,42]. Finally, substances such as the cytokines interferon-gamma and hypoxia-inducible factor-alpha were equally involved in the EMT [43,44].
miR-145-5p is expected to be a key target for the treatment of nasal polyps. The current treatments for nasal polyps include corticosteroid-based medications and surgery when conservative treatment fails, and the emergence of biologics such as dupilumab offers a new treatment approach [45]. In recent years, the development of gene therapy has brought new perspectives to the treatment of disease. Short RNA therapies, such as miRNAs and siRNAs, can perform therapeutic functions without altering the genetic information of the host DNA [46]. In this study, we revealed the strong therapeutic potential of miR-145-5p in nasal polyps. Therefore, in the future, the targeted delivery of miR-145-5p in nasal polyp tissues by means of peptide modification and nanotechnology is expected to be a new therapeutic tool [47] that would effectively solve the drawbacks such as the likelihood of recurrence and many surgical risks caused by the current traditional means for nasal polyp treatment.
Although we substantiated our hypothesis through experimental findings and previous research, this study still exhibits certain limitations. First, our study was limited to in vitro experiments and lacked in vivo experiments and nasal polyp animal models. As a result, we were unable to investigate the details of the mechanism underlying the association between miR-145-5p and nasal polyp formation in greater depth. Specifically, we have not undertaken a more in-depth exploration of the mechanisms through which miR-145-5p may be implicated in the EMT via alternative pathways.
HIGHLIGHTS
▪ The formation of nasal polyps in patients with chronic rhinosinusitis is closely related to epithelial-mesenchymal transition (EMT).
▪ miR-145-5p was down-regulated in nasal polyps and elevated miR-145-5p levels could reverse transforming growth factor-β1-induced EMT in primary human nasal epithelial cells.
▪ miR-145-5p emerges as a promising target to inhibit nasal polyp formation. Provide a theoretical basis for nanoparticle-mediated miR-145-5p delivery for treating nasal polyps.
Notes
No potential conflict of interest relevant to this article was reported.
AUTHOR CONTRIBUTIONS
Conceptualization: XLL, DCG, MYL. Methodology: WTW, YQY. Validation: DCG, MYL. Formal analysis: MYZ, XLP. Investigation: MYZ, XLP, XLL. Resources: FX. Data curation: XLP, WTW, YQY. Visualization: MYZ, MYL. Supervision: DCG, MYL. Project administration: DCG, MYL. Funding acquisition: MYL. Writing–original draft: MYZ. Writing–review & editing: all authors.
Acknowledgements
This work was supported by the Suzhou Clinical Medical Center of Otorhinolaryngology, Head and Neck Surgery (SZLCYXZX 202102), and Suzhou People’s Livelihood Science and Technology Project (SYS2020115).
The pattern diagrams in this work were created with BioRender.com.
SUPPLEMENTARY MATERIALS
Supplementary materials can be found online at https://doi.org/10.21053/ceo.2023.00025.
Transforming growth factor (TGF)-β1 expression was elevated in nasal polyps. (A) Western blot analysis for protein expression of TGF-β1 in normal tissues (N1, N2, N3) and nasal polyps (P1, P2, P3). (B) ImageJ was used to calculate the relative expression rate. (C) Quantitative real-time polymerase chain reaction for mRNA expression of TGF-β1 in normal tissues and nasal polyp tissues. All data are shown as mean±standard deviation. n=3 per group. **P<0.01.